Mar 2, 2020 · 11 min read
Weak Supervision for Science and Medicine: A Year in Review
Jared Dunnmon and Chris Ré. Referencing work by other members of Hazy Research.
While we are proud of the adoption that weak supervision has seen in industry, we are just as excited about the impact it can have in science and medicine. This post reviews recent work that leverages weak supervision to provide real-world value to scientists and clinicians.
Over the last four years, we have seen a paradigm shift in which machine learning models traditionally trained using expensive hand-labeled data have been increasingly replaced by those built using massive, weakly supervised training sets powered by systems like Snorkel that draw on principles from Data Programming and Software 2.0. Weak supervision has dramatically reduced the time and effort required to create training datasets for deep machine learning models -- often from person-years to person-hours! -- by combining massive amounts of unlabeled data with noisy heuristics provided by domain experts and unsupervised generative modeling techniques.1
While we are proud of the adoption that weak supervision techniques have seen in industry (e.g. Snorkel Drybell @ Google, Overton @ Apple, Osprey @ Intel), we are just as excited about the impact they have begun to have in science and medicine. Below we provide a brief review of recent work2 that leverages weak supervision to provide real-world value to scientists and clinicians, focusing on three major modes of use that we have observed: (1) single-modality applications of weak supervision, (2) weak supervision using signal drawn from the byproducts -- or exhaust -- of existing workflows, and (3) cross-modal weak supervision, where information from one modality is used to supervise a model built over another.
We are both excited and humbled by the advances that weak supervision has yielded in science and medicine thus far, and are incredibly thankful for the engagement of the many physicians and domain scientists with whom we have collaborated -- none of the work described below would be possible without their efforts. We look forward to working even more closely with these communities in the future, and to seeing continued growth at the intersection of weakly supervised machine learning and scientific discovery in the years ahead!
Single-Modality Applications of Weak Supervision
A natural approach to applying weak supervision to problems in science and medicine is to identify use cases where human processes are slow or expensive, and to subsequently build weakly supervised machine learning models that can perform those same processes at improved speed or scale. We highlight three of these applications here that have leveraged the data programming technique implemented in Snorkel: extracting patient outcomes from clinical notes for medical device surveillance, screening for rare cardiac abnormalities on high-dimensional volumetric imaging, and building a database of Genome Wide Association Studies (GWAS) from the biomedical literature.
Improving Medical Device Surveillance for Hip Implants
Post-market medical device surveillance is challenging for manufacturers, regulatory agencies, and healthcare providers alike because information relevant to tracking patient outcomes over time is fractured across clinical notes and structured records. Using hip replacements -- one of the most common implantable devices -- as a test case, Callahan et al. used Snorkel to build weakly supervised machine learning models that accurately extracted implant details and reports of complications and pain from electronic health records with up to 96.3% precision, 98.5% recall, and 97.4% F1; improved classification performance by 12.8 -- 53.9% over rule-based methods; and detected over six times as many complication events compared to using structured data alone. Using these additional events to assess complication-free survivorship of different implant systems, they found significant variation between implants, including for risk of revision surgery, which could not be detected using structured data alone. These weakly supervised models offer an exciting opportunity to create scalable solutions for national medical device surveillance using electronic health records.
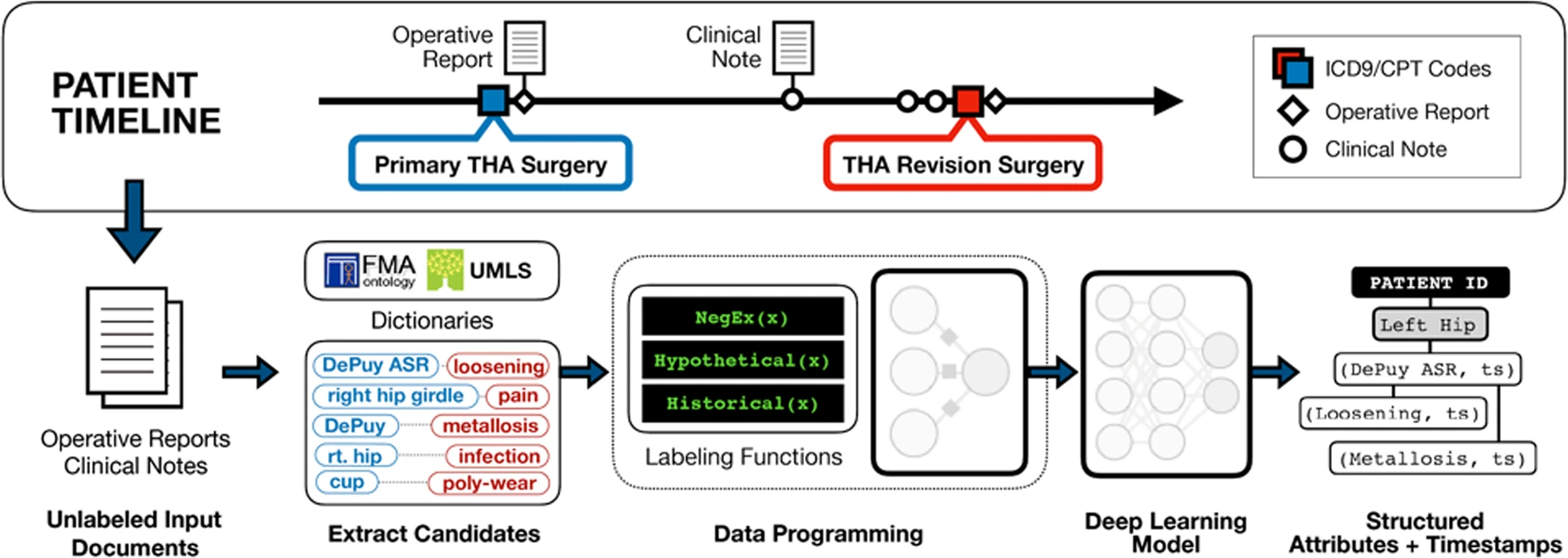
Figure 1: Weak supervision pipeline for medical device surveillance from Callahan et al. Note that weak supervision heuristics -- or labeling functions -- are written directly over the raw text, and that the models are used to extract outcomes from text reports.
Screening for Rare Cardiac Abnormalities on MRI
Biomedical repositories such as the UK Biobank provide increasing access to prospectively collected cardiac imaging. However, these data are unlabeled, which creates barriers to their use in supervised machine learning. Fries et al. have recently developed a weakly supervised deep learning model for classification of aortic valve malformations by applying Snorkel to over 4,000 unlabeled cardiac MRI sequences. Not only was this model able to substantially outperform a supervised model trained on hand-labeled MRIs, but in an orthogonal validation experiment using health outcomes data, it was able to identify individuals with a 1.8-fold increase in risk of a major adverse cardiac event. The success of this study demonstrates the potential of weakly supervised models to increase access to high-throughput screening that would be infeasible using human analysis alone.
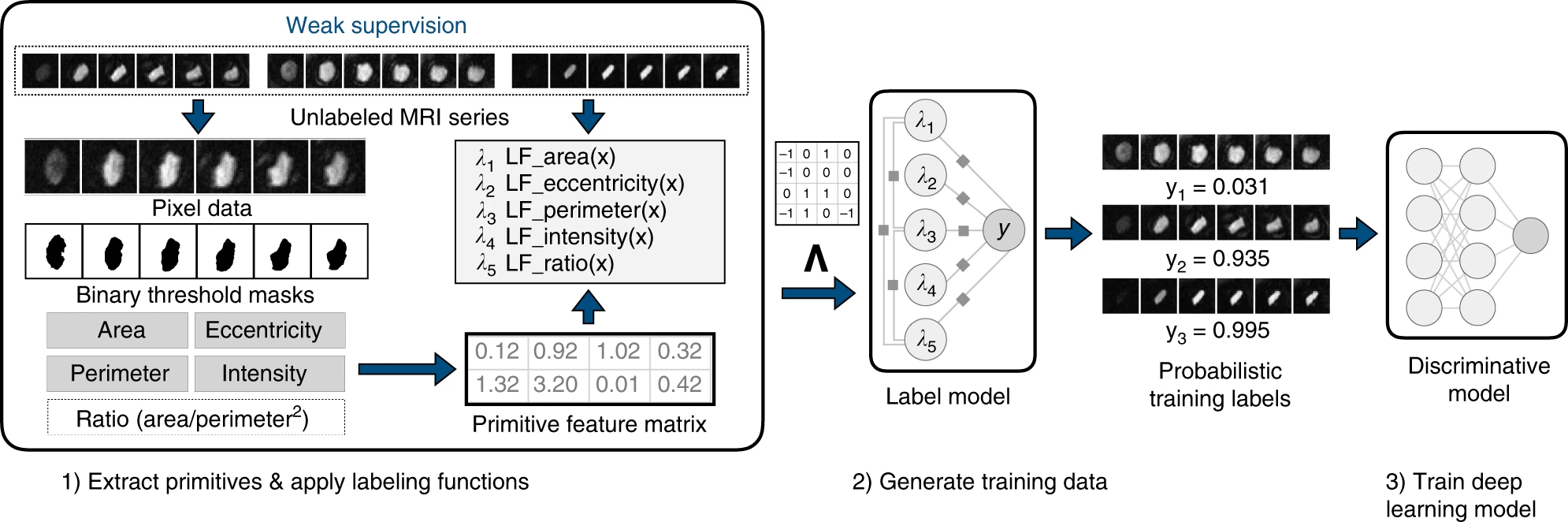
Figure 2: Weak supervision pipeline for aortic abnormality detection from Fries et al. Note that in this case, labeling functions are written directly over primitives (area, perimeter, etc.) computed using an unsupervised segmentation of the valve, meaning that only information from the image was used to create weak labels.
Building a Machine-Compiled Database of Genome-Wide Association Studies
Tens of thousands of genotype-phenotype associations have been discovered to date, yet not all of them are easily accessible to scientists. Kuleshov et al. describe GWASkb, a machine-compiled knowledge base of genetic associations collected from the scientific literature using automated information extraction algorithms. Their information extraction system -- built using Snorkel -- is able to assist database curators by automatically collecting over 6,000 associations from open-access publications with an estimated recall of 60 -- 80% and with an estimated precision of 78 -- 94% (measured relative to existing manually curated knowledge bases). Importantly, the machine-curated database took only six months to build, while its human-curated equivalent took nearly a decade. This work represents a promising step towards making the curation of scientific literature for downstream analysis more efficient using automated systems.
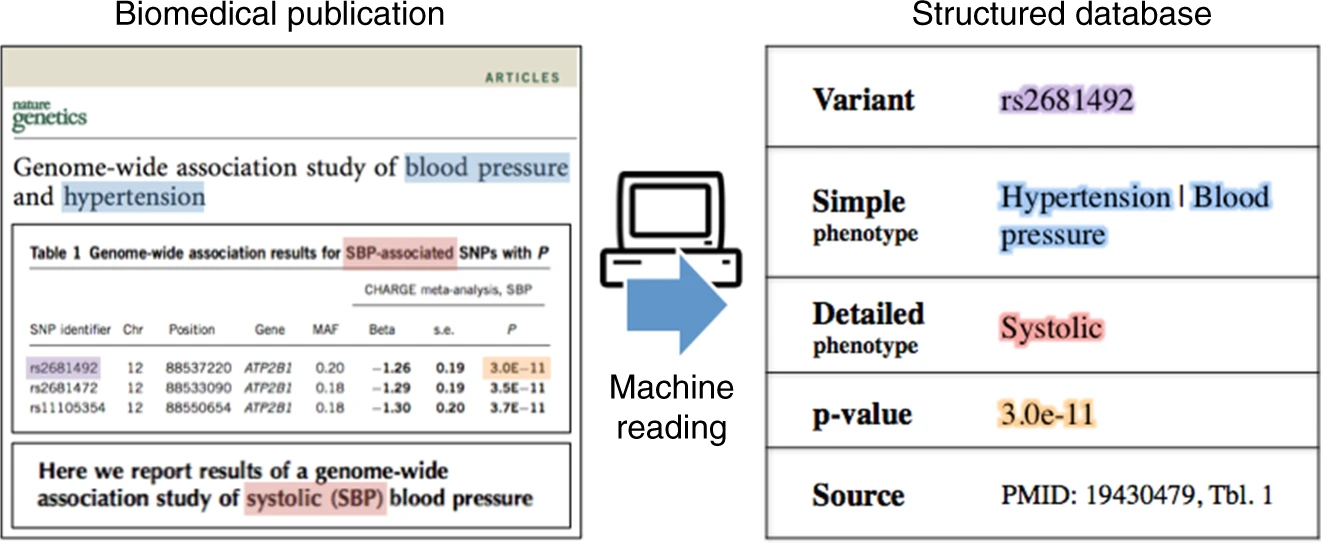
Figure 3. Diagram of the types of data compiled from the literature by Kuleshov et al. using weakly supervised extraction models built in Snorkel.
Weak Supervision Using Workflow Exhaust
In addition to heuristics composed over raw data, signals appropriate for weak supervision can also be generated directly from workflow exhaust such as clinical metadata or upstream analysis processes. We highlight two use cases from medicine that directly demonstrate the clinical value of using workflow exhaust to weakly supervise machine learning models: triage of chest radiographs and seizure detection on electroencephalography (EEG).
Supervising Automated Radiograph Triage Models with Clinical Summary Codes
The work of Dunnmon et al. was one the first to demonstrate that weakly supervised machine learning models could provide clinically useful performance on chest radiograph triage tasks. Specifically, instead of using hand-labeled data, these researchers pulled noisy supervision signal from a set of clinical summary codes recorded in the course of clinical practice. In addition to demonstrating state-of-the-art performance on this task, they found that high levels of performance could be achieved even with relatively modest numbers of weakly labeled datapoints (on the order of 20,000).
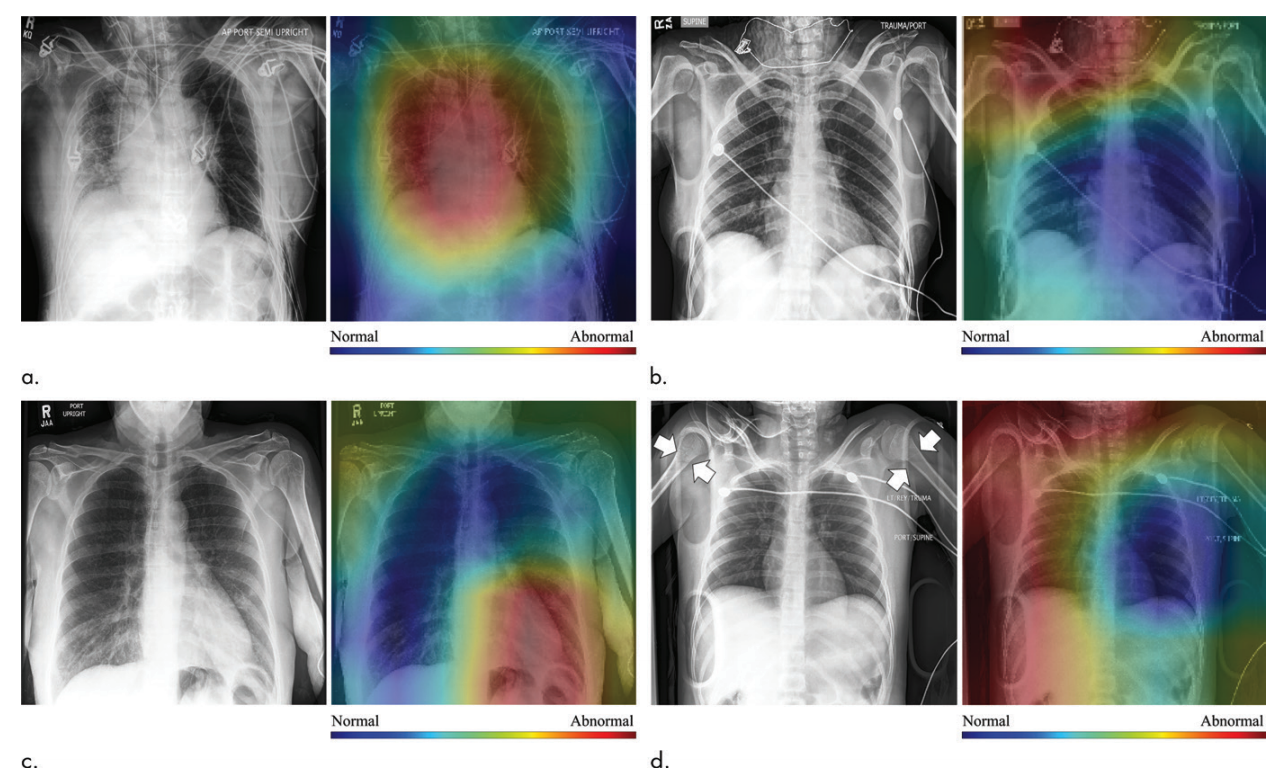
Figure 4: True positive (top left), false positive (top right), false negative (bottom left), and true negative (bottom right) examples from the chest radiograph triage study of Dunnmon et al.; see the paper for more details.
Supervising Seizure Detection Models with Technician Annotations
In a similar vein, recent work by Saab et al. shows that state-of-the-art seizure detection models for EEG can be trained using weak annotations obtained within the course of usual clinical practice. While a neurologist always provides the final determination of whether a patient has experienced a seizure, EEG technicians generally annotate areas to which the physician should pay attention before the physician reads the study. Saab et al. show that this technician signal -- which is both low-precision and low-recall in aggregate -- can nonetheless be used as weak supervision for seizure detection models that outperform currently deployed clinical systems.
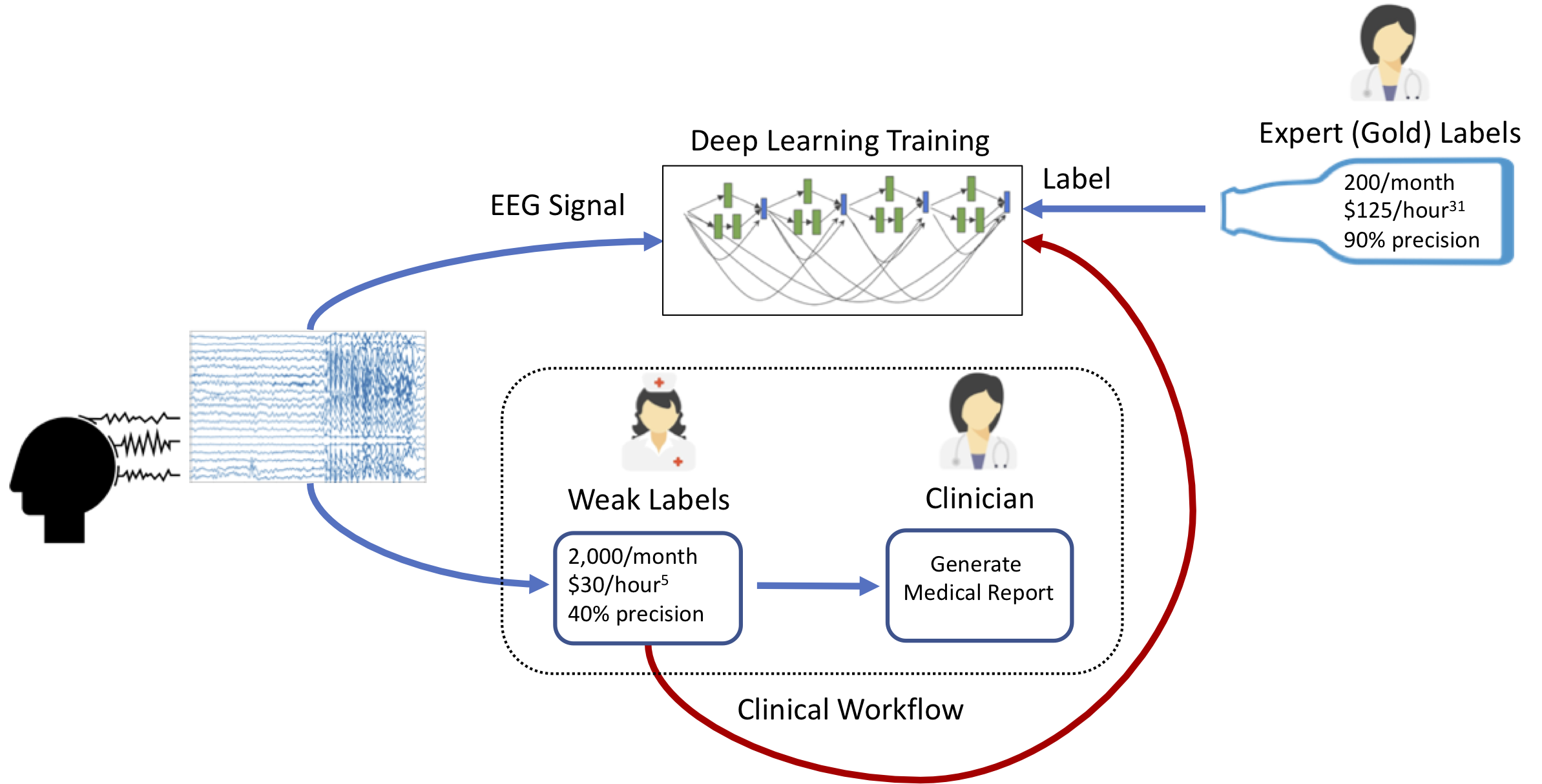
Figure 5: Annotation workflow from Saab et al.; note that the inexpensive, relatively imprecise weak labels sourced from EEG technicians can be used directly as supervision for a highly performant seizure detection system.
Cross-Modal Weak Supervision
Finally, it is often the case that data from a given modality (e.g. imaging) is unlabeled, but that there exists another modality (e.g. unstructured text reports) that contains useful supervision signal. This concept -- which we call cross-modal weak supervision -- has shown particular value in clinical workflows, where application of data programming in a cross modal setting has reduced the time required to build useful machine learning models by an order of magnitude across medical use cases spanning radiograph triage, hemorrhage detection on computed tomography, and automated seizure detection on EEG. We highlight the work of Dunnmon et al., which makes critical first steps in demonstrating the importance of this technique across each of these applications.
Enabling Rapid Medical Machine Learning with Cross-Modal Data Programming
Development of machine learning models for medical diagnostics is often limited by a lack of labeled data. Further, many modalities do not admit a useful unsupervised featurization over which heuristics can be written, making single-modality weak supervision in the style of Fries et al. difficult. Dunnmon et al. solve this problem by observing that real-world clinical data rarely exist in a vacuum, and that the text reports accompanying clinical diagnostic studies often contain the information required to provide supervision for clinically useful machine learning tasks. They extend the usual data programming paradigm by first creating weak training labels over the text using Snorkel, and then training a Long Short-Term Memory (LSTM) network that maps the raw text to these weak labels even on examples where all heuristics abstain. The output of this LSTM is used to train downstream classification models for diagnostic studies. Importantly, Dunnmon et al. show that models trained in this way are often statistically indistinguishable from those trained using an equivalent amount of hand-labeled data, that weakly supervised model performance scales similarly with unlabeled data to how fully supervised models do with labeled data, and that weak supervision provided an average 93% reduction in required labeling resources with respect to hand labeling to achieve equivalent performance.
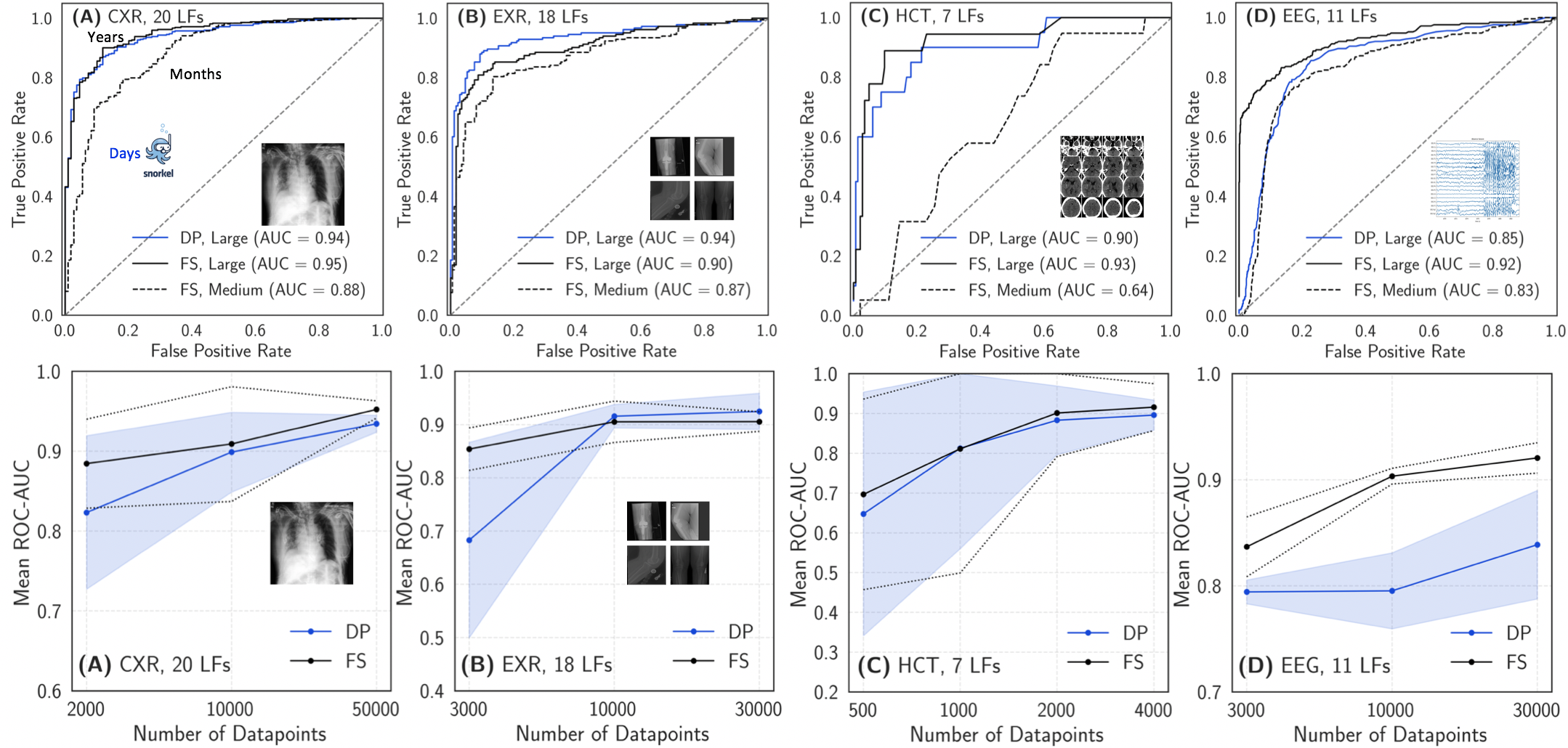
Figure 6: Results from Dunnmon et al. showing (top) the absolute performance of hand-labeled (black) versus weakly supervised (blue) models and (bottom) scaling behavior of both model types. (A) is single chest radiograph triage, (B) is extremity radiograph series triage, (C) is hemorrhage detection on head CT, and (D) is seizure detection on EEG.
Extracting Fine-Grained Output from Coarse-Grained Annotations
As followup to the study of Dunnmon et al., Saab et al. evaluated the degree to which coarse-grained cross-modal weak supervision could be combined with informed architecture specification choices to create models that yield clinically useful fine-grained information even when only trained with coarse-grained information. Concretely, cross-modal weak supervision was used to extract training labels for a Intracranial Hemorrhage (ICH) detection task on computed tomography of the head, and an attention-based multiple instance learning model was used to classify each scan. When the attention weights and in-plane occlusion maps were examined, the data demonstrated that the 2-D axial slices that were given high attention weights were exactly those which contained the hemorrhage, and that occlusion maps yielded large values at spatial regions within the brain that overlapped with a radiologist segmentation of the hemorrhage. Importantly, these results suggest that machine learning models weakly supervised using coarse-grained labels can still provide fine-grained information that is extremely useful to clinicians.
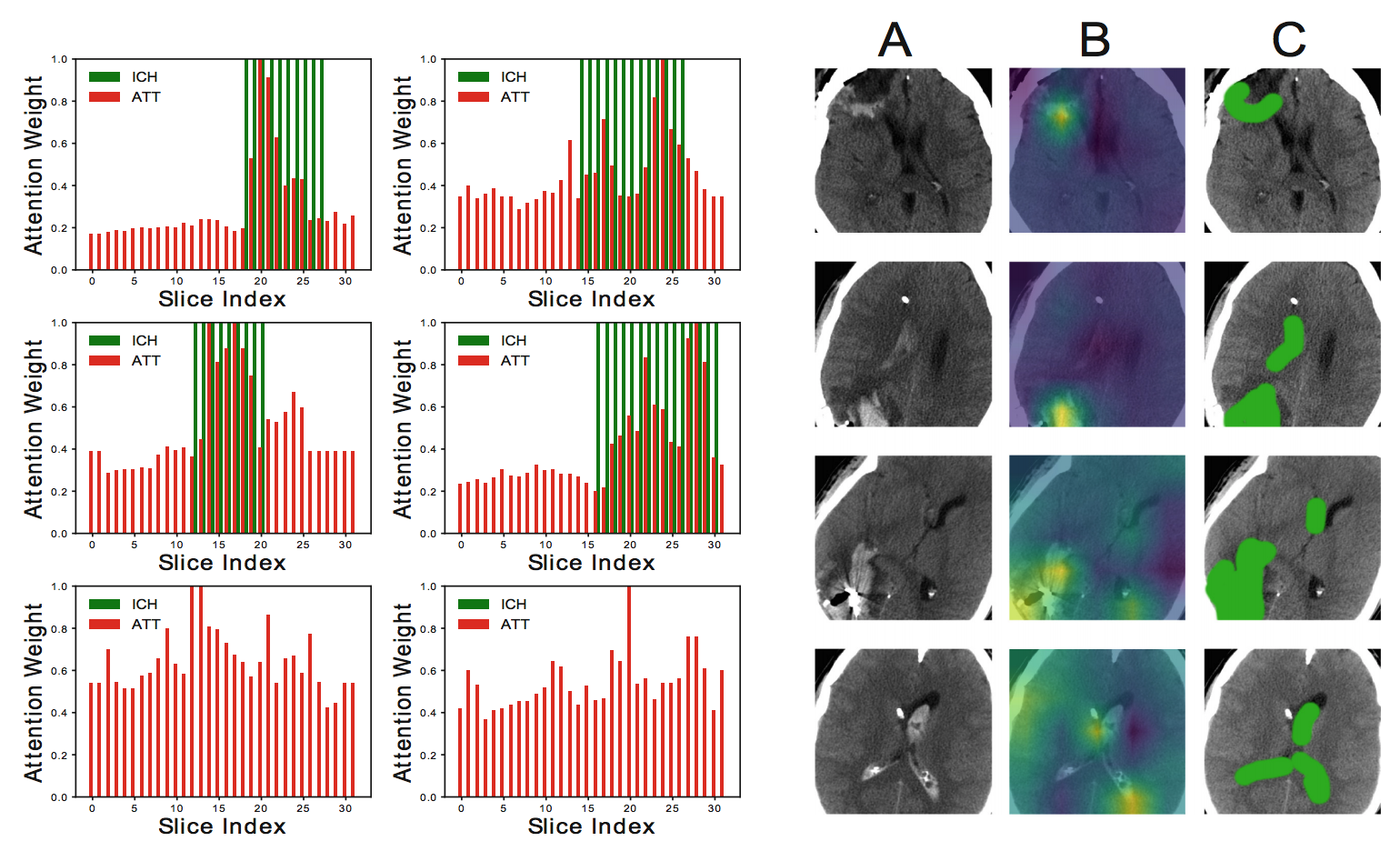
Figure 7: Results from Saab et al. on showing (left) normalized attention weights ("ATT", red) compared to ground-truth slice annotations for ICH ("ICH", green), where the bottom two cases have no ICH and (right) ICH in raw CT slices (column A) as a hyperdense (i.e. whiter) substance as compared within the intracranial vault to the surrounding parenchyma and fluid; voxel-level hand labels (column C) coarsely identify voxels most likely to represent ICH, while occlusion maps (column B) demonstrate favorable correlation with the coarsely labeled voxels.
Into the Future
We are excited to keep exploring applications of weak supervision in science and medicine, and see several distinct paths for future work in this area. The first of these is moving from active, declarative supervision -- such as having domain experts provide heuristics for data programming -- to passively collected observational supervision, where human activity can be directly used to provide training signal. The second of these is using approaches from weak supervision, data augmentation, and robust optimization to handle real-world problems such as hidden stratification, where models perform poorly on unlabeled subsets that are semantically meaningful and practically important.
More detail on these ideas and more can be found on our group's website -- please get in touch if these ideas excite you as much as they excite us!